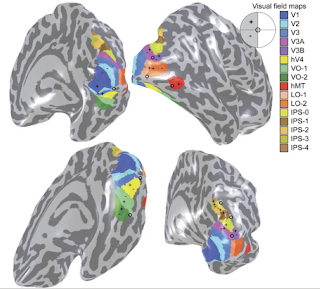
Retinotopy is the topographical relation between visual field and neuronal organization in visual cortex. You may get deep insight if you check Larsson et al.2006 and Wandell et al. 2007 .
There are two main functional scans to determine retinotopic visual areas;
one is rotating Wedges for polar angle organization and the other one is the Rings for eccentric information. Wedges were performed both in CW and CCW direction and Rings were performed in contracting and expanding direction.
So in routine retinotopy scan procedure, we have following scans;
* t1MPR 176 slice TR:2600 TE:3.02 ms
* ep2d for overlapping wedges ccw 3x3x3 TR 2 s / TE:35ms / F.A.:71 270meas
* ep2d for overlapping wedges cw 3x3x3 TR 2 s / TE:35ms / F.A.:71 270 meas
* ep2d for expanding rings 3x3x3 TR 2 s / TE:35ms / F.A.:71 160meas
* ep2d for contracting rings 3x3x3 TR 2 s / TE:35ms / F.A.:71 160meas
* ep2d for horizontal and vertical wedges 3x3x3 TR 2 s / TE:35ms / F.A.:71 159meas
=====================
1) Create "subject.vmr" file and make and AC-PC alignment
AC point shown with cursor in axial view.
PC point shown with cursor in axial view.
this alignment will create "subject_AC-PC.vmr" and "subject-to-subject_AC-PC.trf" file.
2) Create all functional ".fmr" files and preprocess only the first ".fmr" file with default options.
3) Align first ".fmr" file to "subject.vmr" using 3Dtools FMR-VMR co-registration.
you will get transformation files with following extensions ".IA" stands for initial alignment and ".FA" stands for final alignment.
note: in some conditions final alignment may fail ,you may check the manual alignment box under final alignment card in FMR-VMR co-registration menu. Default is identity matrix which means only initial alignment is done but there is still ".FA" file which has no effect on co-registration. Use this option when you are sure about initial alignment is sufficient, you may overlay .VTC extension and check registration quality with F8 button later on.
4) Open native vmr file, subject.vmr
then go to create 3D-aligned VTC
Create VTC file by using the 4 files created in previous steps;
Select spatial transformation as "To ACPC"
5)Check VTC volume for registration quality;
open AC-PC aligned vmr file and attach VTC file that you've created.
then go to the 3DvolumeTools - Spatial Transformation-Show VTC vol.
cruise your cursor around and select hypo-intense changes like edges of the brain or ventricles. Change volume by clicking F8 and be sure VTC volume is at its desired place.
6) The rest functional scans are not preprocessed and not registered either;
Use intra session alignment option for rest of the FMR preprocessing steps
and leave rest as default.
7) Use same ".IA" , ".FA" and ".TRF" files (that you've created for first functional scan) and use AC_PC.TRF for rest of the volume time courses(VTC).Same as step "4)" just different ".fmr" file for each functional scan.
8)Now all VTC's are created and images are ready for correlation analysis.
All anatomical analysis should be done and you should have meshes to go on.
http://visionumram.blogspot.com/2012/01/segmentation-in-freesurfer-rest-in.html
9) Check mesh files, use following link if you have any errors at your mesh files especially at cortical locations.
http://support.brainvoyager.com/volume-space/32-segmentation-automatic-cortex/147-manual-postcorrection-of-the-automatic-segmentation-result.html
10) Meshes are ready to inflate;
Use mesh morphing with following parameters under Mesh menu.
Apply inflation for both hemispheres , use smoothed versions (RECOSM)
11) Use background and curvature colors under mesh menu;
check two color mode and smooth 3 times .this will enhance the good looking of the gyrus and sulcus boundaries.
Save this file with "_inf" (inflated) suffix.
12) Open an AC_PC aligned vmr file and attach VTC of wedges scan (CCW or CW).
assign polar angles for your stimuli at stimulation protocol.
13) Use linear correlation maps tool to get phase encoded map of polar angles.
for
counter clockwise left visual field (CCW_lvf)
clockwise right visual field (CW_rvf)
use hemodynamic response function(HRF) at 0 degree visual polar angle
for
counter clockwise right visual field (CCW_rvf)
clockwise left visual field (CW_lvf)
use hemodynamic response function(HRF) at 180degree visual polar angle
enable cross correlation and set #of lags to 18.
14)Open an AC_PC aligned vmr file and attach VTC of rings scan (contracting or expanding).
assign 6 eccentricity for your stimuli at stimulation protocol.
15) Use linear correlation maps tool to get eccentricty encoding of visual field.
use hemodynamic response function(HRF) at 1st eccentricity. for both expanding and contracting rings.
enable cross correlation and set #of lags to 30. (5 level for each eccentricity)
16) Open an AC_PC aligned vmr file and attach VTC of horizontal and vertical wedges scan. Create 2 predictor for vertical and horizontal case and make a GLM analysis for that with following protocol,use default overlay(plus for horizontal and vertical)
17) Use the results of the steps "13)" , "15)" , "16)" which are on VMR files , open
your inflated mesh files. and map those results to each hemisphere using "Surface Maps" tools under mesh menu. Each subject should have 5 functional maps for each hemispheres.
At statistics card ; check lag values and use Mapping_lh_rh_1.olt as LUT.
18) After all .smp files are created for each subject. Now we could define retinotopic borders. In this part only V1,V2 and V3 will be defined.
V1 has lower vertical meridian line at superior
upper vertical meridian line at inferior portion
( defined as green and red color)
V2 has horizontal meridian line both at superior (V2D-V3D) and inferior portion (V2V-V3V) defined as yellow and blue color
V3 visual fields has similar borders like V1
lower vertical meridian line at superior (V3D border)
upper vertical meridian line at inferior portion(V3V border)
defined as green and red color.
19) To draw borders manually , under painting and cutting click "deletion mode"
then click "draw cut mesh mode" .Create dashed line where you're sure about the borders than complete it to a solid line, to make the changes permanant.
Deactive the "draw cut mesh mode " radio button and
Use background and curvature colors under mesh menu;
check two color mode and click curvature and smooth respectively.Then save your mesh with "_BNDRY" prefix, which stands for retinotopic boundries.
20) If everything was fine you should get a consistent visual borders for each functional maps.